Abstract
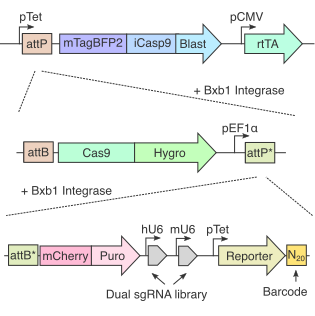
RNAs undergo a complex choreography of metabolic processes that are regulated by thousands of RNA-associated proteins. Here we introduce ReLiC, a scalable and high-throughput RNA-linked CRISPR approach to measure the responses of diverse RNA metabolic processes to knockout of 2,092 human genes encoding all known RNA-associated proteins. ReLiC relies on an iterative strategy to integrate genes encoding Cas9, sgRNAs, and barcoded reporter libraries into a defined genomic locus. Combining ReLiC with polysome fractionation reveals key regulators of ribosome occupancy, uncovering links between translation and proteostasis. Isoform-specific ReLiC captures differential regulation of intron retention and exon skipping by SF3b complex subunits. Chemogenomic ReLiC screens decipher translational regulators upstream of mRNA decay and identify a role for the ribosome collision sensor GCN1 during treatment with the anti-leukemic drug homoharringtonine. Our work demonstrates ReLiC as a powerful framework for discovering and dissecting post-transcriptional regulatory networks in human cells.